The study introduces a safe and effective intratumoral epidermal growth factor receptor (EGFR)-targeted CRISPR-lipid nanoparticle (LNP) delivery strategy for co-encapsulating CRISPR-Cas9 messenger RNA (mRNA) and single guide RNA (sgRNA) to achieve SOX2 gene knockout. We generated functional targeted LNPs (tLNPs) that co-encapsulate Cas9 mRNA with sgRNA targeting SOX2 and are coated with anti-EGFR antibodies. Their uptake, expression in HNSCC cancer cells, and therapeutic effects were evaluated in vitro and in vivo. The results demonstrate that these tLNPs can specifically target tumor cells, induce sustained therapeutic responses, and provide a promising treatment approach for clinically high-recurrence solid tumors such as HNSCC.
1. Experimental Results:
1.1 sgRNA Screening and Gene Editing Efficiency
Three sgRNAs targeting the SOX2 gene (sgSOX2-A, -B, -C) were designed and screened in HNSCC cell lines (UMSCC-104 and FADU), representing different head and neck cancer primary origins. The researchers compared gene editing efficiencies after transfection with Cas9 protein and sgRNAs (forming ribonucleoprotein/RNP complexes) targeting SOX2. Sanger sequencing and Inference of CRISPR Edits (ICE) analysis revealed that sgSOX2-A achieved 58% in vitro gene editing in UMSCC-104 cell line and 40% gene editing in FADU cell line. Consequently, sgSOX2-A was selected as the optimal guide RNA for further in vivo and in vitro experiments.
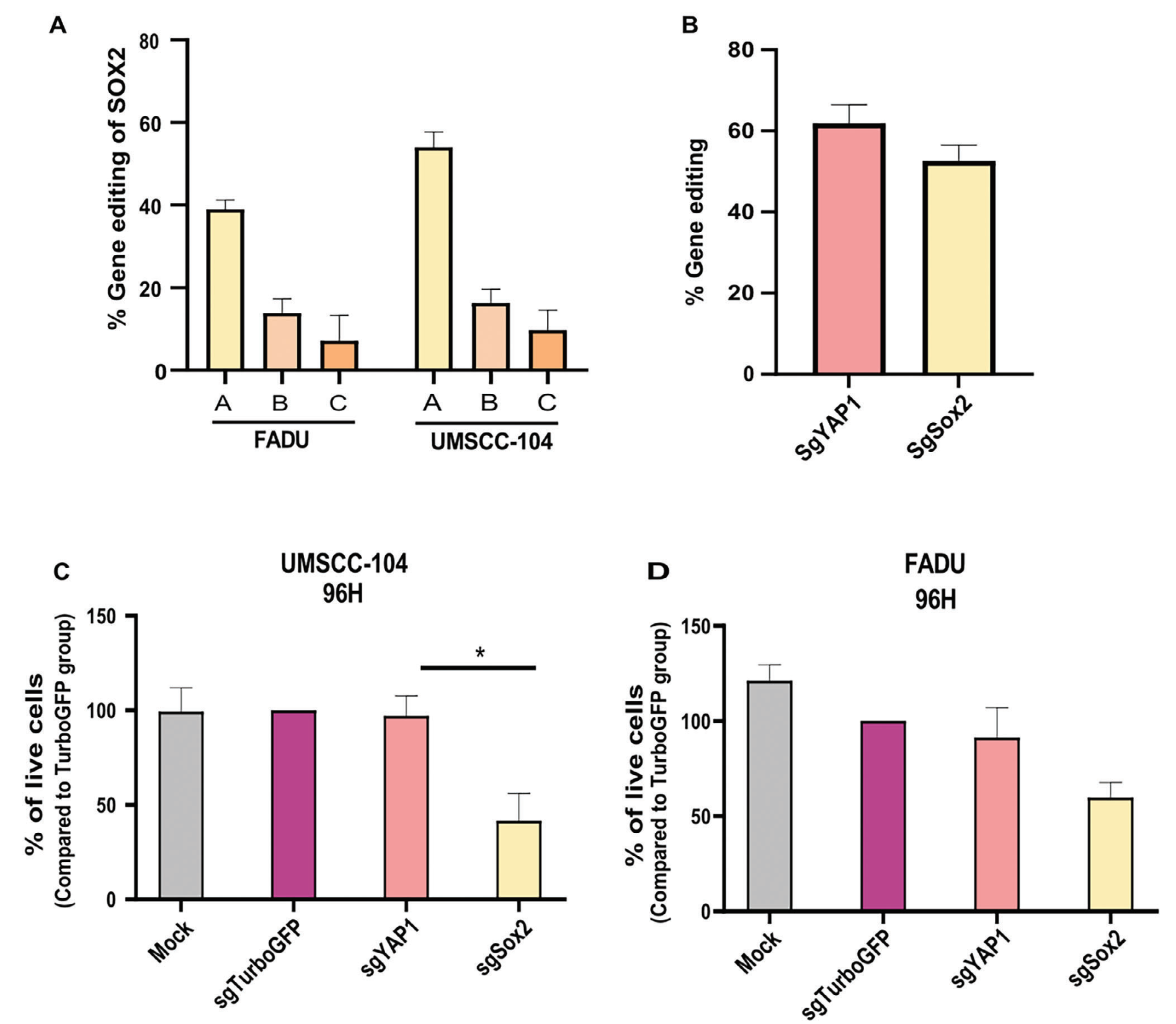
Figure 1: In Vitro sgRNA Screening and Cell Viability Detection
1.2 Impact of Gene Editing on Cell Viability
XTT proliferation assays were performed on UMSCC104 and FADU cells transfected with sgSOX2-ribonucleoprotein (RNP) complexes. Turbo-GFP and YAP1 sgRNA were used as controls. Since YAP1, similar to SOX2, is involved in cancer stem cell characteristics and is overexpressed in HNSCC, it was used as a control for efficient gene editing. Experimental results demonstrated that SOX2 gene editing significantly reduced the viability of HNSCC cells.
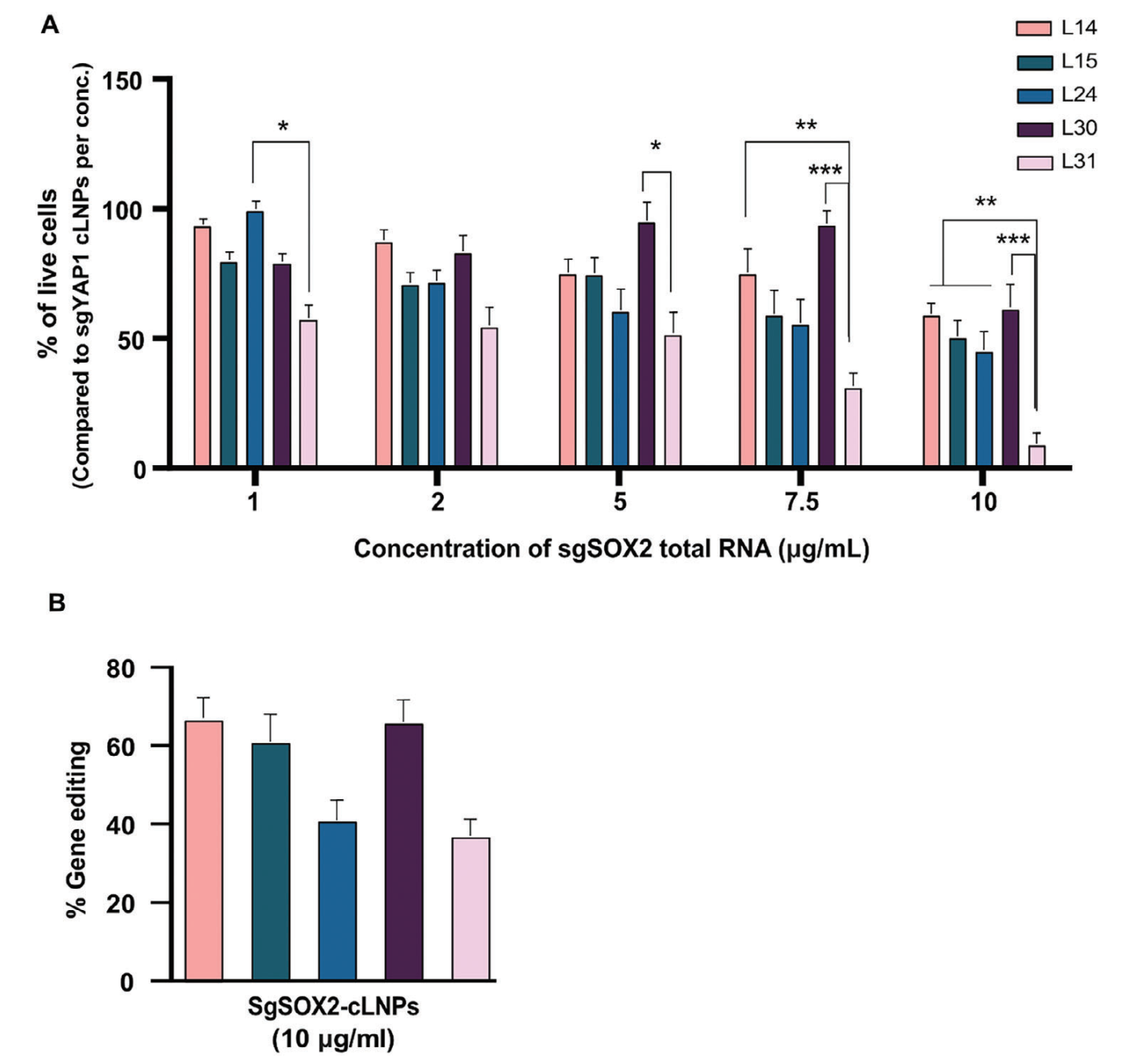
Figure 2: Impact of Gene Editing on Cell Viability and Editing Efficiency Assessment
1.3 Characterization of Targeted LNPs and Gene Editing Efficiency
Targeted lipid nanoparticles (tLNPs) were prepared and characterized by transmission electron microscopy. The results demonstrated that tLNPs possessed an appropriate diameter, low polydispersity index (PDI), and favorable zeta (ζ) potential, indicating good stability and dispersibility. To confirm the gene editing efficiency of tLNPs, gene editing analysis were conducted. The results showed that a single tLNP treatment achieved approximately 17% gene editing at the SOX2 locus.
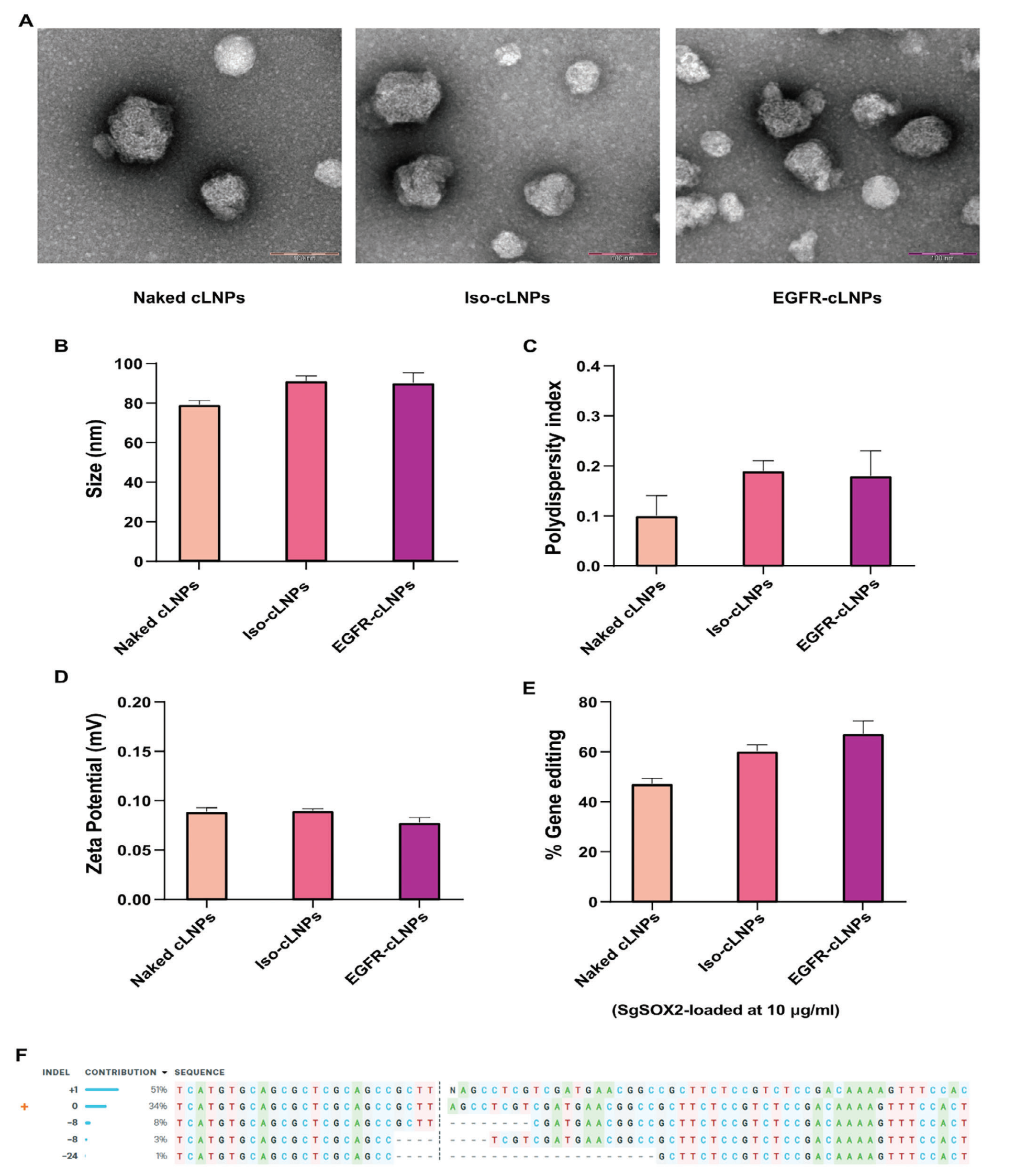
Figure 3: Physicochemical Properties Evaluation and Editing Efficiency of Targeted LNPs
1.4 Biodistribution and Safety Assessment
In vivo studies demonstrated the biodistribution of targeted lipid nanoparticles (tLNPs) specifically target head and neck squamous cell carcinoma (HNSCC) tumors and achieve efficient SOX2 gene knockout. Simultaneously, gene editing percentages in splenic and hepatic cells were analyzed to confirm the absence of off-target editing. The results showed no evidence of off-target editing. Furthermore, immunogenicity and toxicity assessments indicated that SOX2-conjugated LNPs (SOX2-cLNPs) exhibited no toxicity or immunogenicity.
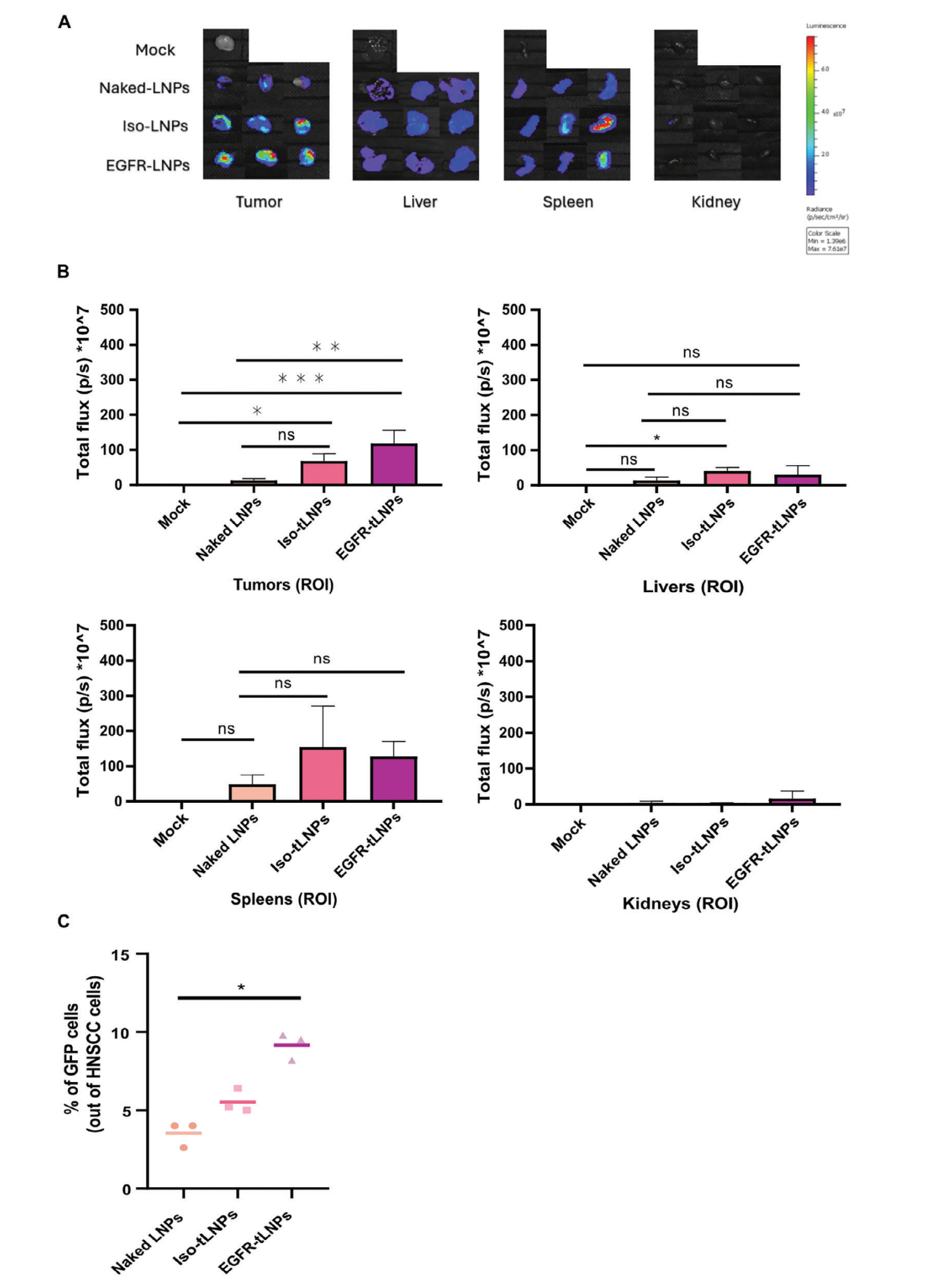
Figure 4: In Vivo Uptake, Distribution, and Expression of Targeted LNPs
1.5 Therapeutic Effects of Targeted tLNPs in HNSCC Mouse Model
To explore the ability of targeted tLNPs to mediate therapeutic gene editing in vivo, their effects on inhibiting head and neck squamous cell carcinoma (HNSCC) tumor growth were evaluated. Using a xenograft HNSCC mouse model, intratumoral injection of targeted tLNPs resulted in 90% tumor growth inhibition and a 90% increase in survival (>84 days) compared to the untreated group. Complete tumor disappearance was observed in 50% of the mice, indicating that tLNPs can efficiently and specifically target HNSCC tumors, achieving sustained therapeutic responses.
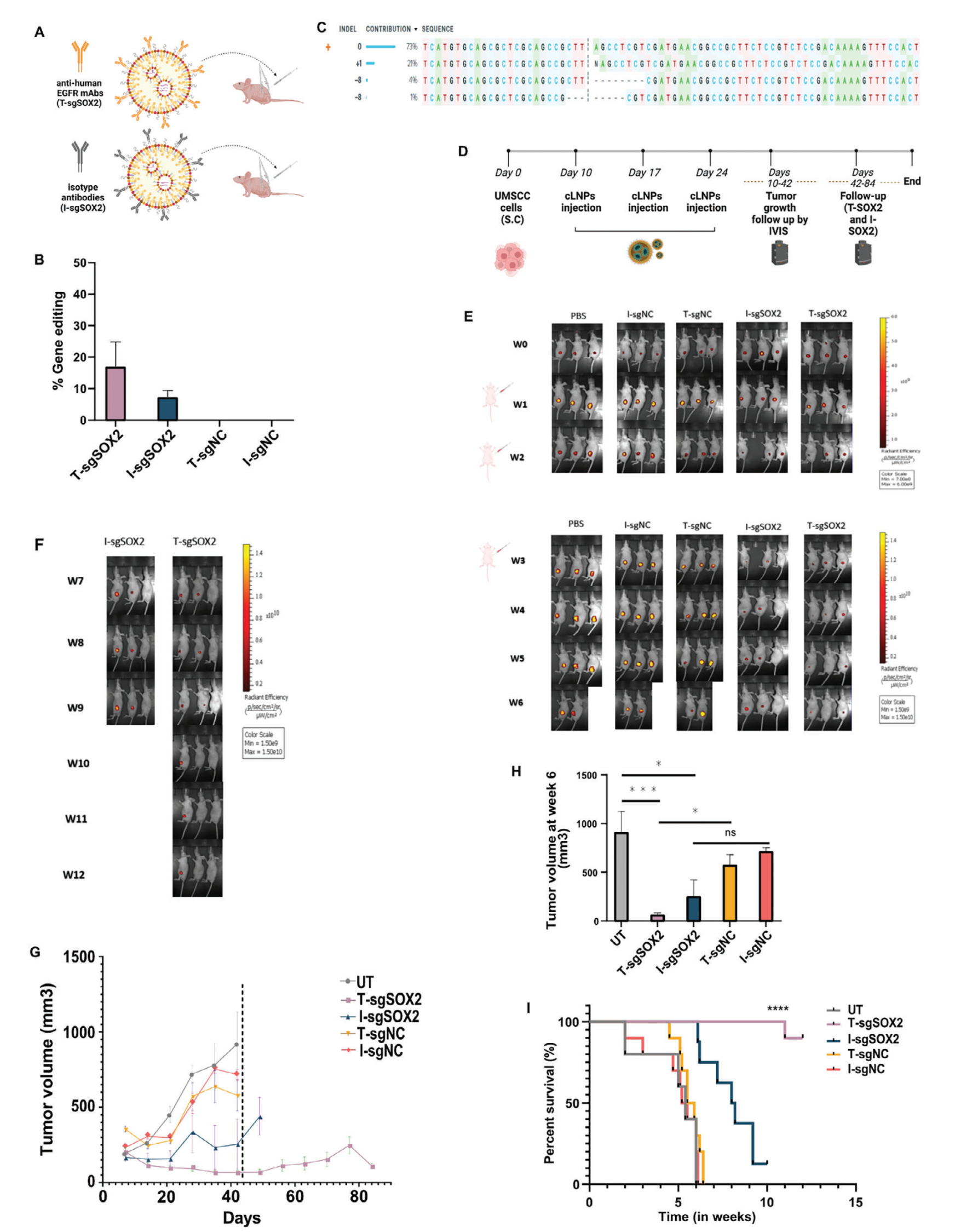
Figure 5: In Vivo Efficacy Evaluation of Targeted LNPs
2. Discussion
This study successfully developed an EGFR-targeted CRISPR- LNP delivery strategy for HNSCC that achieves SOX2 gene knockout. The targeted LNPs (tLNPs) can specifically target tumor cells and induce sustained therapeutic responses, making this strategy promising for the treatment of clinically high-recurrence solid tumors such as HNSCC.
However, the study has several limitations. For instance, although tLNPs exhibited excellent therapeutic effects in HNSCC cancer cells, their applicability in other cancer types requires further validation. Additionally, potential off-target effects of the CRISPR-Cas9 system remain a critical concern. Future research will focus on optimizing tLNP preparation and delivery strategies to further reduce off-target effects and enhance therapeutic efficiency.
3. Conclusion
An EGFR-targeted CRISPR-LNP delivery system for head and neck cancer was successfully developed and its therapeutic potential evaluated. Results showed that these tLNPs can specifically target tumor cells and induce sustained therapeutic responses. This approach offers a promising treatment strategy for clinically high-recurrence solid tumors, warranting further research and potential clinical translation.
References:Masarwy, Razan, et al. "Targeted CRISPR/Cas9 lipid nanoparticles elicits therapeutic genome editing in Head and Neck Cancer." Advanced Science (2024): 2411032.
|